|
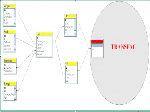
We have developed a relational database system which is aiming at providing a comprehensive overview on all expression sources.
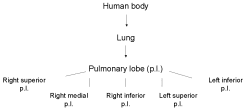
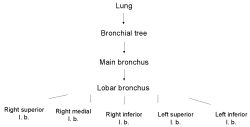
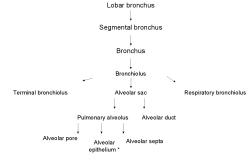
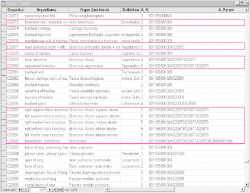
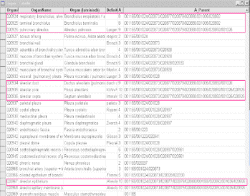
Gene expression sources are organs, tissues, cell types and developmental stages. Therefore CYTOMER is a database of physiological systems (Ssystem-table), developmental stages (Sstage-table), anatomical
structures and substructures (Sorgan-table) and the constituting cell-types (Scell-table). The central table of CYTOMER
is Hub which is a list that links entries of four other tables. The Hub-table represents anatomical / histological knowledge about which cells occur with what kind of function in which organs and at what stages.
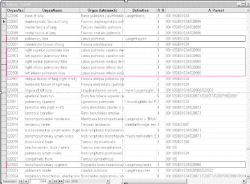
The CYTOMER database is applied to map expression patterns as the TRANSFAC database gives. Therefore entries of the Hub-table have been linked with human transcription factor entries in the
TRANSFAC factor table: 1) CP (Cytomer positive)-column for those expression sources where a certain factor has been shown to be expressed in and 2) CN (Cytomer negative)-column for those expression sources where no
evidence of a certain factor has been published.
CYTOMER is first of all a TRANSFAC-complementing module which enables proper representation of expression patterns. However, CYTOMER is
going to be extended to an independent database system which provides customers with specific aims. |