|
 |
|
General features of Pax |
 |
|
|
Ø |
large family of developmental regulatory genes that encode nuclear transcription factors |
Ø |
early expressed during embryogenesis |
Ø |
normally downregulated in adult stages |
Ø |
involved in organogenesis (mainly CNS) |
Ø |
role in morphological boundaries and early regionalisation |
Ø |
proto-oncogenes |
|
|
 |
 |
|
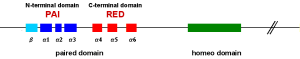
DNA binding of Pax proteins |
 |
 |
|
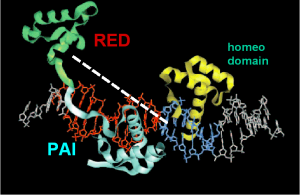
The DNA binding is mainly confirmed by helix 3 of the PAI domain in the major groove whereas helix 1+2 are antiparallel
(figure 3). Additionally the homeo and RED domain can be involved so various combinations of DNA binding are possible (Jun et al. 1996). |
 |
 |
|
Evolution of Pax |
 |
 |
|
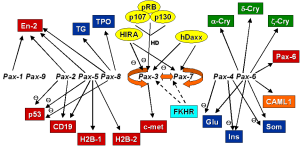
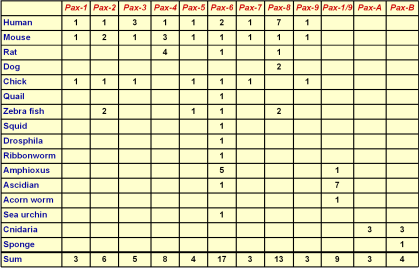
According to sequence similarity of the paired box and the existence of a homeobox and octapeptide, Pax genes were grouped
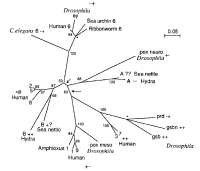
into four paralog goups (table 1). Surprisingly in more primitive organisms certain precursors corresponding to these groups were found (Sun et al. 1997, figure 4). This indicates that gene duplication events lead to
divergation of more members within one group based on one ancestral Pax gene. |
|
|
 |
|
|
 |
|
|
Figure 4: phyllogenetic tree of Pax genes based on paired box similarities (Sun et al. 1997)
|
|
|