| Contents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
ReactionA reaction in TRANSPATH® is a term for all kinds of interactions between signaling entities (molecule or gene) in signaling or regulatory events. The character of the interaction is more closely defined in the effect field by a set of terms. Reactions as processes are not physical entities like molecules, yet they are the central point in a signal transduction database. By representing these reactions between molecules as separate nodes in the graph, it becomes possible to store their properties and annotate them. Since many reactions in signal transduction are catalyzed, and most catalyzed reactions are quasi-unidirectional, all reactions stored in the database are by default unidirectional. Equilibrium reactions are identified by the term "reversible" in the effect field. As of release 5.1, there are five kinds of reactions in the database: semantic, indirect, pathway step, decomposition, and molecular evidence. These terms are stored in the type field
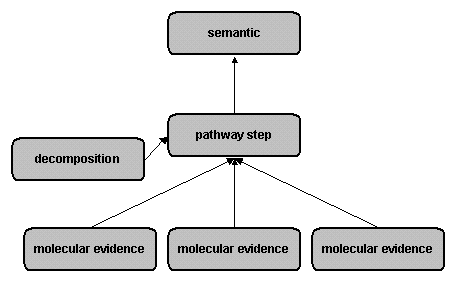
Reaction hierarchyThe reaction hierarchy has been introduced in release 5.1 to connect the patchwork of molecular evidence reactions (different species and modified forms in the various experiments) to a mechanistic level that models consecutive pathways. This pathway level consists of pathway step, auxiliary decomposition and indirect reactions (in cases where the direct mechanism for a part of the pathway is not known). In combination with reaction chains, this level is used to model canonical pathways and networks. The semantic projection level gives a broader shortcut overview of the signal transduction pathways without biochemical details (Fig. 2)
Mechanistic reactions: molecular evidence, pathway step, decompositionThe classical chemical reaction notation
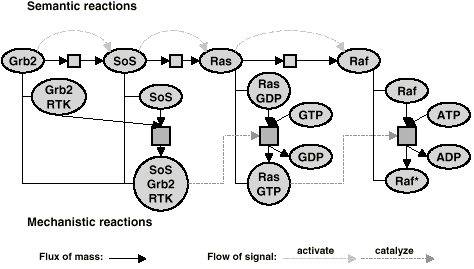
can model aggregation into a complex, dissociation of a complex, chemical modification and catalysis in the detail necessary to capture data from the primary literature. An example for this kind of data is given in Fig.3.
We call this representation mechanistic, because it reflects how the molecules interact, and not how the signal is transported. It is well-established and familiar and is used for the pathway step, decomposition and molecular evidence reaction types. Molecules act as enzymes, substrates, modulators, or products, and any reaction has exactly one or no enzyme, one or more substrates, any number of modulators, either inhibitors or activators, and one or more products. There is semantic information in assigning some of the inputs to the enzyme or modulator collections, which tells us something about the local meaning of the molecule for the reaction. This view demands a detailed knowledge of the mechanisms of transduction. It makes no assumptions about the molecule states. All the different states of a molecule have to be listed as separate entities. Semantic reactionsThe representation of reactions commonly encountered in scientific review papers can be seen in Fig. 4. We call this representation semantic, because it assigns a meaning to the states of the molecules for the overall network, "active" or "inactive". It is easy to understand and it is familiar to scientists from the papers. It shows how the signal flows through the network. The reactions need additional information, that is if they activate or inhibit the target molecule. The reactions in this scheme are binary. Molecules act as signal donors or signal acceptors to the reactions.
This view implicitly assumes that each molecule exists in an active and an inactive state. It is not necessary to differentiate between them, as it is understood that incoming activating reactions always refer to the inactive state, incoming inactivating reactions and outgoing reactions refer to the active state. Because the states/modified forms are implied for semantic reactions, we link them to basic molecule entries in the database. Saying that a molecule has an "active" or "inactive" state is a semantic statement. Both states undergo reactions, and the decision as to which is which can consequently only be determined in the larger context of the whole network. TranslocationsIn a translocation, the same molecule enters and leaves the reaction with a changed spatial context. This process takes some time, which is important for the dynamic behavior of the network. Translocations cannot be represented in the basic mechanistic model which assumes all reaction partners are present in the same reaction space. Since a molecule is just associated with a list of locations, we cannot differentiate between, for example, the cytosolic and the nuclear form of the molecule. A reaction that moves a molecule from one form to the other is then a loopback from the molecule to itself. Therefore we associated the two locations with the connection entries between the molecule and the reaction. The term translocation is assigned in the effect field. More detailed information about the data model of TRANSPATH can be found at: "Consistent re-modeling of signaling pathways and its implementation in the TRANSPATH database" Genome Inf. Ser. 15, 244-254. [Pubmed] [.pdf] |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||